Genomics analysis is playing a key role in COVID-19 studies, as the data from sequencing projects is helping researchers better understand and characterize the coronavirus.
With NVIDIA Parabricks, researchers can integrate GPU power into existing genomics workflows and rapidly sequence fragments to enhance research.
In the COMPUTE4COVID webinar series, we discuss in detail how sequencing workflows are important for COVID research, and how GPUs are helping accelerate these workflows.
Sequencing for COVID-19
There are three major workflows important to understanding the novel virus:
- Viral Genome sequencing
- Human RNA-sequencing from infected cells
- Human genome association studies
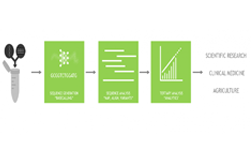
Before workflows can begin, DNA needs to be broken down into fragments and sent through a sequencer. There are two lengths of DNA fragments that can be sequenced: short reads, which consist of 100 base pairs; and long reads, which consist of 10,000 base pairs.
These fragments are put through sequencers and recombined into the original DNA strand. Through this process researchers can read the human genome and viral genome.
Researchers studying the coronavirus must continuously sequence the viral genomer to understand how the virus mutates over time. This information can then be usedin developing a vaccine that is effective against all strains of the virus.
When researchers sequence fragments of DNA, the next step is to align and assemble the genome.
If there is a reference genome they can align to, then the researchers have an idea of what the DNA should look like, and they can compare sequences to find small variances between them.
However, sometimes the researchers don’t have a reference genome to align to. In this case, they need to take the DNA fragments and decipher in which order they should be assembled. Both workflows are computationally intensive, but with GPUs, they can be significantly enhanced.
NVIDIA Parabricks Speeds Genomic Analysis
Assembling a genome is not only time-consuming, but requires vast computational resources. These workflows used to be done on CPUs, but now with NVIDIA Parabricks, researchers can harness the power of GPUs to accelerate existing algorithms and speed up analysis.
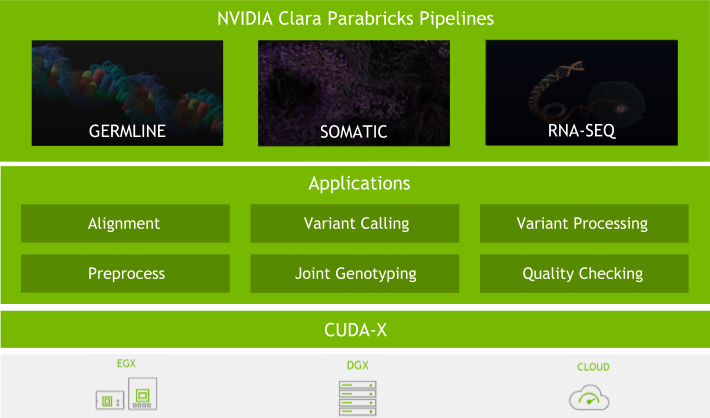
Parabricks is a suite of GPU-accelerated software for analysis of data from short-read sequencing technology. Built on the CUDA-X platform, Parabricks can be deployed on-premise or in the cloud.
Parabricks provides three relevant offerings:
- BWA (Burrows-Wheeler Aligner) for short read alignment to a reference genome
- STAR (Spliced Transcripts Alignment to a Reference) for short read alignment to a reference genome. Specifically for RNA sequencing.
- GATK (Genome Analysis Toolkit) for variant calling from short-read human genome sequencing
With Parabricks, researchers can accelerate COVID research by leveraging GPUs for computing, using single nodes to run an entire analysis, and significantly reduce the overall cost of computing.
Learn more about NVIDIA Parabricks and see how other companies are using technology to battle the coronavirus in the COMPUTE4COVID webinar series.
More Resources
To learn more about how AI and accelerated computing are helping researchers and developers fight the pandemic, visit our COVID-19 Research Hub.