The Human Genome Center (HGC) at University of Tokyo announced a new genomics platform to accelerate genomic analysis by 40X compared to a CPU-based environment, utilizing NVIDIA Clara Parabricks Pipelines genomics software. The genomic analysis will run on SHIROKANE, HGC’s fastest supercomputer for life sciences in Japan, and powered by NVIDIA DGX A100. The platform will be available to users on April 1, 2021. SHIROKANE helps researchers quickly process massive amounts of genomic data and is incredibly powerful with many nodes, a capacity of over 400 TFLOPS, and a storage capacity of over 12PB. The ultimate goal of analyzing so much genomic data is to glean insights about germline and somatic variants to move closer to precision medicine.
Today, patients are prescribed medicines that work for the majority of people, but are often ineffective as they are not tailored to a specific patient’s genetic profile. Precision medicine aims to provide more specific therapeutics for patients, utilizing information from whole genome sequencing and other clinical data. As a national strategy, Japan’s Ministry of Health, Labor and Welfare formulated the Execution Plan for Whole Genome Analysis in December 2019, to focus on the areas of cancer and intractable diseases. The plan will take up to 3 years, aims to sequence 92,000 patients, and will ultimately help create a database that will be utilized by research institutions, pharmaceutical companies, and university hospitals for drug development and disease prevention.
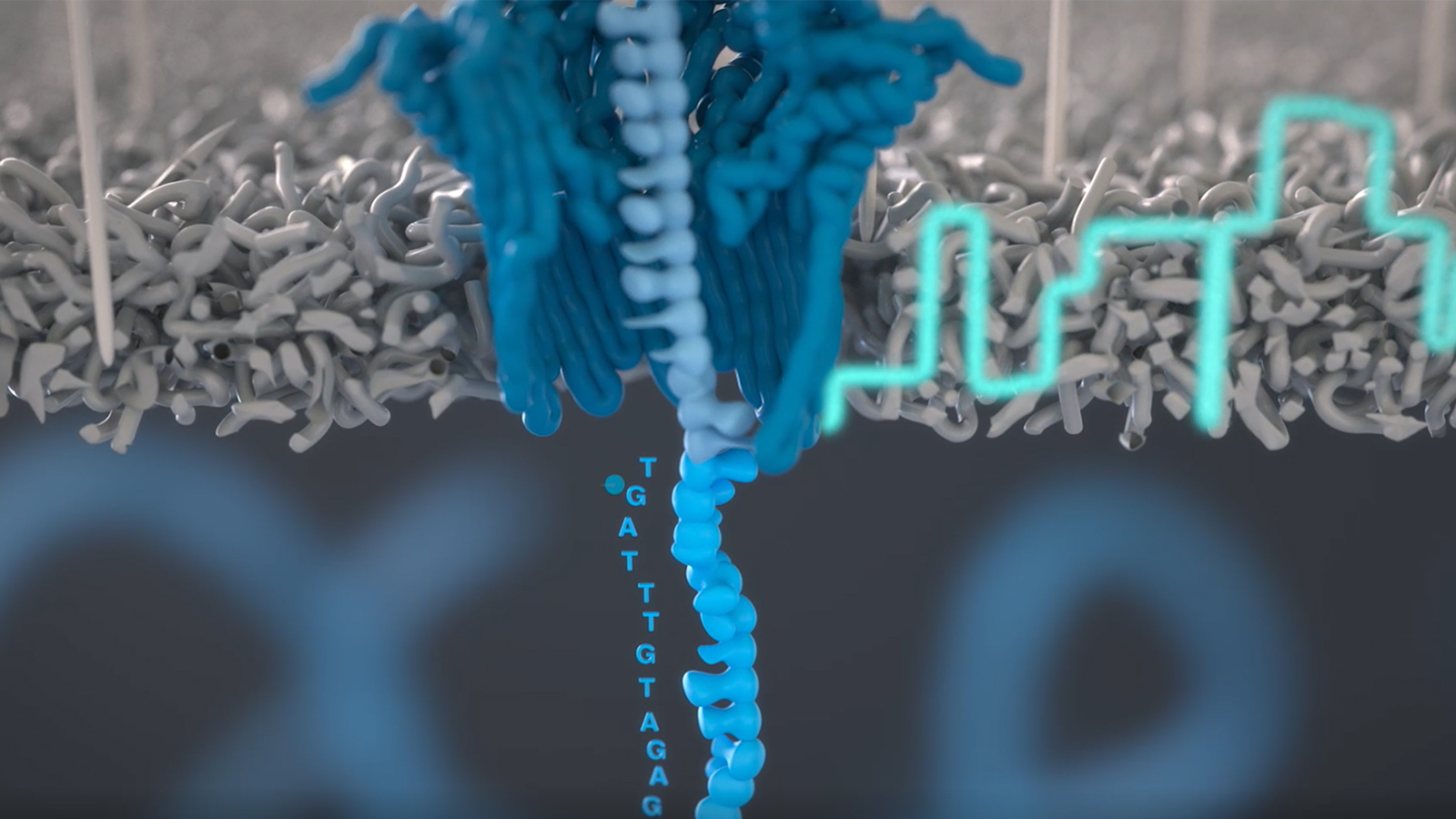
Whole genome sequencing (WGS) has been widely recognized for its comprehensive analysis, and its increasing usefulness in areas such as infectious diseases and cancer. WGS examines the complete DNA of an organism while exome sequencing examines the protein coding regions or genes, which make up about 1.5% of the human genome. WGS requires several times the sequence depth and can be done quickly with accelerated genomic analysis, like with NVIDIA CLARA Parabricks Pipelines.
Professor Seiya Imoto, Director of HGC said, “The Human Genome Analysis Center at the Institute of Medical Science has been working on refining whole-genome data analysis and shortening the analysis time in cancer genomic medicine. This time, we evaluated Parabricks for implementation on all GPU servers on SHIROKANE. Its high speed and functions are indispensable for the future of large-scale whole-genome analysis. The whole-genome data analysis capability is equivalent to hundreds of conventional CPU servers and was implemented on the GPU server. We will realize a state-of-the-art high-speed whole-genome data analysis environment that greatly accelerates genome research for SHIROKANE users.”
Clara Parabricks Pipelines’ accelerates genomic analysis by utilizing the parallel computing performance of GPUs. Many germline and somatic callers have been accelerated in Clara Parabricks Pipelines including Google’s DeepVariant, which identifies genome variants in sequencing data using convolutional neural networks (CNN). Previously, whole genome analysis typically would take 20 hours or more per sample in a general CPU environment, however on SHIROKANE, powered by NVIDIA DGX A100s GPUs, the analysis takes less than 30 minutes. HGC put Parabricks Pipelines in production on 16 of the 80xNVIDIA V100 GPUs installed on SHIROKANE in February 2020 and is open to users from life science companies.
The genomic analysis proved to be faster than expected, and with the increasing number of users accessing SHIROKANE, there was a need to further super power SHIROKANE. Eight NVIDIA DGX A100 systems were recently added in 2021 to SHIROKANE, for a total of 88xGPU servers coupled with Parabricks Pipelines to accelerate large-scale genomic workloads. In addition, SHIROKANE provides free access to researchers working on SARS-CoV-2, in an effort to expedite insights about the virus and those infected by the virus. A joint research group called “The Corona Suppression Task Force” formulated at HGC will consist of experts from seven universities and research institutions to focus on various new coronavirus infections.

NVIDIA DGX A100 is the universal system for all AI workloads, offering unprecedented compute density, performance, and flexibility in the world’s first 5 petaFLOPS AI system. It features NVIDIA A100 Tensor Core GPUs, enabling customers to consolidate training, inference and analytics into a unified, easy to deploy infrastructure.
“NVIDIA has been investing for several years in anticipation of the coming era of large-scale whole-genome analysis,” commented Masataka Osaki, NVIDIA Japan Country Manager and VP Corporate Sales. “The greatest achievement, Parabricks, along with the latest DGX A100 system, is greatly helping Japan’s premier cancer genome research center. NVIDIA’s platform will be the foundation that supports whole-genome research in Japan, and it is expected that the elucidation of genes associated with cancer and intractable diseases will progress dramatically.”
Professor Seiya Imoto, Director of the Human Genome Analysis Center, at the University of Tokyo, is presenting a talk on whole genome analysis at the GTC21 conference April 12-16, which is free this year. Register here.