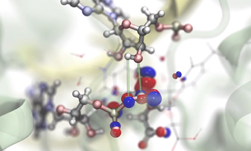
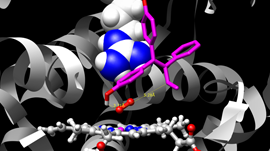
Erik Lindahl, Professor of Biophysics at Stockholm University, talks about using the GPU-accelerated GROMACS application to simulate protein dynamics. This approach helps researchers learn how to design better drugs, combat alcoholism and understand how certain diseases occur.
Lindahl mentions they started using CUDA in their work nearly five years ago and now 90% of their computational resources are GPUs. “It’s everything we do.”
Learn more about the GROMACS molecular simulation toolkit at GROMACS.org.
Share your GPU-accelerated science with us at http://nvda.ly/Vpjxr
Watch more scientists and researchers share how accelerated computing is #thepathforward: Watch Now
Share Your Science: Unraveling Membrane Proteins with GPUs
Feb 09, 2016
Discuss (0)
Related resources
- GTC session: Poster Reception (Sponsored by Cadence)
- GTC session: Accelerating Drug Discovery: Optimizing Dynamic GPU Workflows with CUDA Graphs, Mapped Memory, C++ Coroutines, and More
- GTC session: Enhancing Discovery in TechBio: Leveraging AI for Enzyme and Drug Discovery
- Webinar: Bringing Drugs to Clinics Faster with NVIDIA Computing
- Webinar: Transforming Molecular Design
- Webinar: Accelerating Large-Scale Genomics Research