By Dong Yang, Thomas Sanford, and Daguang Xu.
Last year, the National Institutes of Health (NIH) and NVIDIA began working together on the development of several clinical deep learning tools in diagnostic and interventional imaging. The research involved the analysis of prostate cancer using imaging and pathology data from clinical trials at the NIH.
Prostate cancer is a significant contributor of cancer mortality in the USA, with around 31,000 deaths estimated this year alone.
Traditional methods of prostate cancer diagnosis include a series of tests: digital rectal exam (DRE) and serum prostate-specific antigen (PSA) followed by a transrectal ultrasound (TRUS) guided needle biopsy. Multiparametric MRI (mpMRI) of the prostate has been shown to improve the ability to diagnose clinically significant cancer better than standard methods in multiple studies, but can be more time consuming for physicians.
To help physicians in this effort, NVIDIA and the NIH set out to develop a tool that could be used to better assess both the physical and the cellular presentation of the disease.
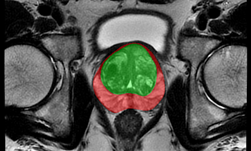
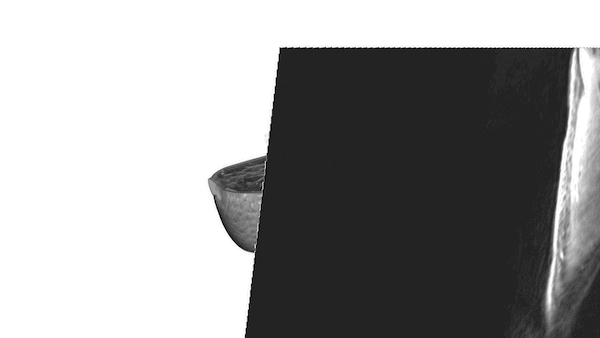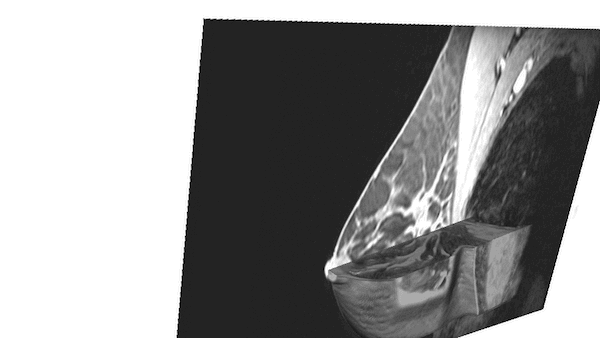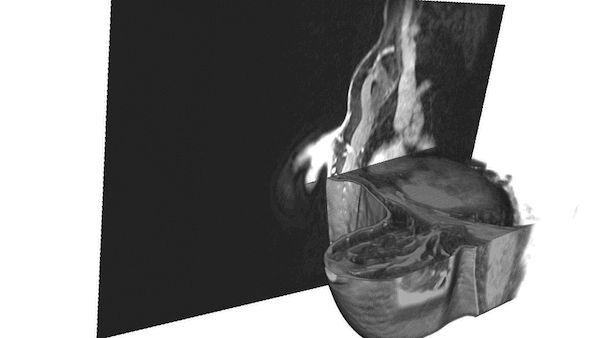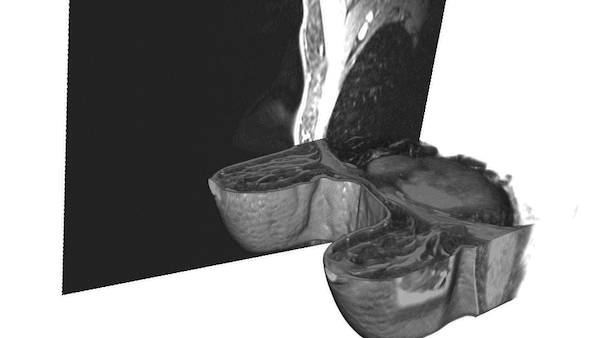
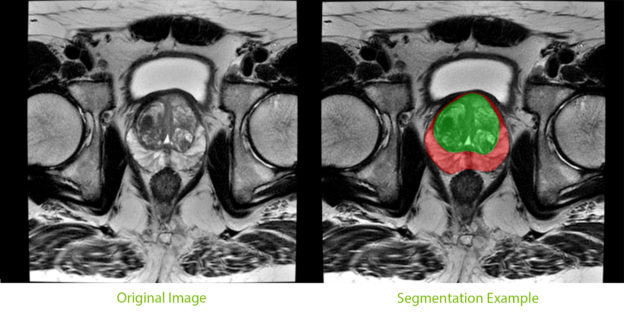
An important component of the tool, segmentation of the prostate from surrounding tissue on MRI images, is useful for a variety of clinical purposes. Segmentation allows for accurate determination of prostate volume and prostate-specific antigen (PSA) density. Both of these measurements can inform the course of treatment for a patient. Segmentation also allows for registration of MRI with other modalities such as ultrasound and PET, and image-guided biopsy and therapy, enabling more accurate targeting of treatment or biopsies.
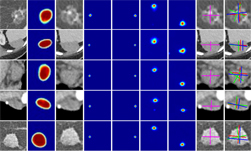
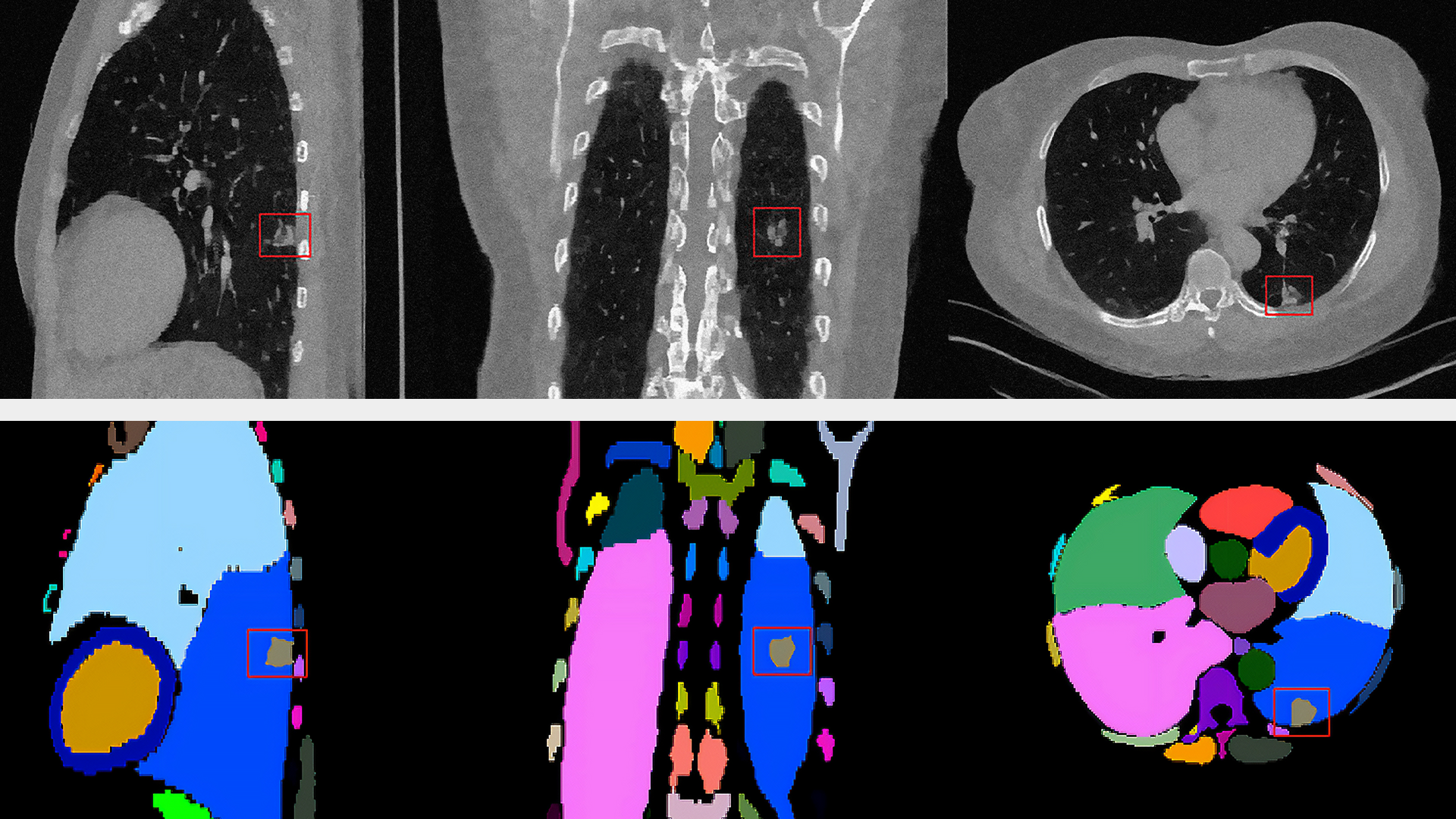
NIH radiologists collected 465 mpMRI data from multiple medical centers, representing multiple MRI vendors (i.e. Siemens, Philips, GE), and various center-specific MRI protocols. The prostate boundaries were manually traced in three planes on T2-weighted MRI by a radiologist with over 10 years of experience in prostate MRI. NVIDIA’s scientists then using the Clara Train SDK, developed a 3D deep learning-based pipeline based on a previous workflow that had won the runner-up award in the “Medical Segmentation Decathlon 2018”. A hybrid 2D-3D neural network was used, and the model was evaluated on 98 patients of unseen data. This method achieved a DICE score of 0.922. In contrast, the DICE score between different radiologists’ annotations is 0.919. Therefore, the team concluded that this approach achieves similar performance to the highly experienced radiologists that annotated the data.
There are usually large variations between the data from different MRI scans due to several factors such as different scanning protocols, scanner vendors, patient populations, etc. Thus, models trained on data from the source domain usually degrades dramatically on other data from unseen domains. In this project, we developed a domain generalization method based on the augmentation transforms which are part of Clara Train framework. No data or annotations from the test domain were used in training or fine tuning.
The model was trained using data provided by NIH, then was locked and evaluated on multiple prostate MRI datasets from public challenges (PROMISE12, NCI-ISBI2013, ProstateX, medical segmentation decathlon, etc). It achieved comparable or better performance versus the state-of-the-art algorithms on every single dataset. On average, it achieved a DICE score of 0.894 versus 0.904 of state-of-the-art algorithms. The state-of-the-art algorithms were trained and evaluated using the data from the same domain while our workflow did not see the data or annotations in the test domain during training.
We implemented and trained the approach using NVIDIA V100 GPUs with the TensorFlow.
The joint effort has now resulted in a best-in-class model that can potentially help improve diagnostic and treatment accuracy for prostate cancer patients.
The end-to-end workflow and networks we discuss here have already been included in the Clara Train SDK. The model trained using medical segmentation decathlon data is already available for download from NGC.
We plan to release the model trained using the NIH dataset in a future version of the Clara Train SDK.
About the authors
Dong Yang, PhD (NVIDIA Researcher)
Dong Yang is an applied research scientist at AI-Infra of NVIDIA. He is specialized in medical image processing, and he is currently working on crafting deep learning approaches to solving particular medical imaging problems, with the goal of improving and inventing new & effective clinical workflows.
Thomas Sanford, MD (NIH):
Dr Sanford is a urologic surgeon specializing cancers of the genitourinary system. He is currently a fellow in the Molecular Imaging Program at the National Institutes of Health working on improving diagnosis of prostate cancer using deep learning.
Daguang Xu, PhD (NVIDIA Researcher)
Daguang Xu is now a research manager at AI-Infra of NVIDIA. He is leading a research team in healthcare AI, focusing on developing world-class machine learning and deep learning-based methods to solve the challenging problems in medical domain.