‘Meet the Researcher’ is a new series in which we spotlight different researchers in academia who are using GPUs to accelerate their work. This month we spotlight Rommie Amaro, professor and endowed chair in the Department of Chemistry and Biochemistry at the University of California, San Diego.
Amaro is also the principal investigator of the Amaro Lab, which is broadly concerned with the development and application of state-of-the-art computational and theoretical techniques to investigate the structure, function, and dynamics of complex biological systems for applications to drug discovery.
This year, she led a team of 28 researchers at a number of different institutions, combining high performance computing (HPC) and AI to provide the clearest view to date of the coronavirus, winning a special Gordon Bell Prize in November 2020.
What are your research areas of focus?
I like to call myself a computational biophysical chemist. In general, we use computational models to explore biological and chemical systems. The idea of being able to use mathematics and computing to understand how biological systems work is just fascinating to me. Over the years, we’ve applied this to areas such as cancer and DNA editing.
What motivated you to pursue your recent research area of focus in supercomputing and the fight against COVID?
We had been working for a number of years on influenza virus, and so after 7-8 years of work on that, we published a paper in February on simulating the influenza virus envelope. That project was really a labor of love. It presented, for the first time, molecular simulations of a lipid enveloped virus in atomic detail. The size was 160 million atoms, very large, and to get to that point we had to overcome multiple technical challenges to simulating viruses in such detail.
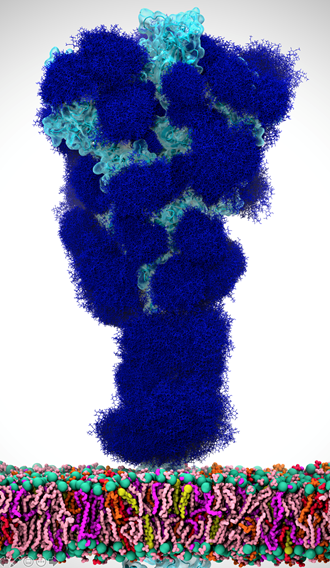
When SARS-CoV-2 hit, it was an immediate and natural pivot to focus on COVID. As soon as the McLellan group deposited the spike structure in the bioRxiv, we scooped up the data and started working with it, aiming to simulate the complete spike structure in all its atomic detail.
What problems does your research address?
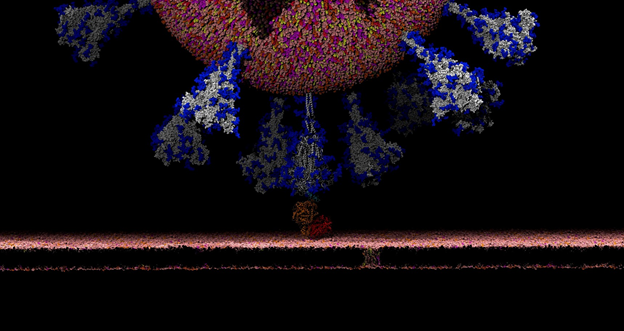
There’s this aspect of the virus that’s very intriguing. Viruses in general have evolved something called a glycan shield, which is basically a sugary coating that the virus uses to mask itself from the human immune system.
All of the cells in our body are coated with different types of sugar molecules, called glycans. The viruses have evolved this way of looking just like other human cells by covering themselves in the same soft of sugary coating. That way, when viruses get into our bodies, our immune system doesn’t see spike protein, it sees this sugary coating. Since it looks like any other cell, the human body doesn’t recognize it as a threat and therefore doesn’t activate the immune system response.
Experimentally, we know these glycans are there but can’t take pictures of them because the sugars move around quite a bit, one can’t get a crisp image of what they actually look like. This is where computing has a lot to give because we can basically create an atomic level structure of what those sugar molecules look like and then that gives us the picture of the sugar structures and what they’re doing. That’s what our research addresses.
Our simulations gave people the first views of what the glycan shield looks like on the viral spike protein. For researchers, it’s not only knowing where that shield is, but it’s even more critical knowing where the shield is not because there are holes in the shield, vulnerabilities of the virus. We can use that information to understand where and how antibodies bind and how to use this information in the design of novel therapeutics.
What is the (expected) impact of your work on the field/community/world?
The reason that we care about simulating the virus in all its atomic detail is that it helps us understand how the virus works, the mechanisms of viral infection, as well as to understand and design new therapeutics. For example, drug molecules that target different virus protein molecules or different parts of the virus. It also helps us understand how antibodies work, their design with vaccines. Having information about where all the atoms are and what they’re doing is really critical.
How have you used NVIDIA technology in your current research?
The main tool my team is using is molecular dynamics simulations. We like to think of this tool as a computational microscope. We take data from different sources, bring these data streams together to create these highly detailed, accurate models of the virus.
The simulations run on chips like NVIDIA GPU systems, and use supercomputing sites that use NVIDIA GPUs. The kind of math these simulations require ports super well to GPUs. That’s where NVIDIA technology has been transformative: it’s allowed us to speed up our calculations tremendously. It’s basically making our microscope really powerful–getting better, more accurate views of the virus much faster. The sooner we have that information, the sooner we can develop drugs and therapeutics to fight the virus.
What were some of the challenges you faced while pursuing your research?
This whole year has been extraordinary. It’s a really crazy time to be a scientist, in the sense of being on the front lines trying to understand the mysteries of COVID-19. There’s so much data coming in from all over the world. We’ve had so many sleepless nights since February/March, working around the clock.
At the same time, there’s been so many amazing collaborations. So many people around the world focused on one single problem. This doesn’t happen, typically we’re each working on different things.
What’s next for your research?
We’re just at the beginning, there’s so much we want to do. Now that we’ve got the virus built and simulated, and that was honestly an incredible feat in terms of how fast we were able to do it, the next is to build and simulate the host cell, the human cell. We want to understand how the virus latches on because it’s a pretty complex process.
Another aspect I’m really excited to study—there are a lot of questions about how and why the virus is airborne transmissible. One of the things we’re planning to do next is to build aerosol particles that have the virus inside of them. These systems will have about one billion atoms. This will help us understand what’s happening to the virus in the air and how long it lives and how it can infect people under a range of conditions, such as relative humidity.
There’s just so much to do.
Any advice for new researchers, or early career professionals, especially to those who are inspired and motivated by your work?
Find your passion. It won’t necessarily be science for everybody. We need so many kinds of people to make the world go round.
Find that thing that lights you up, that you just want to keep coming back to. When I was at university and Saturday came around, everyone would go tailgating. I looked forward to going into the lab. When you really find your passion, your work doesn’t feel like your job. And when your passion unites with the ability to serve others or a greater good, it’s so incredibly rewarding.
Also, education is super important–stay at it, stay in it, to get a better life.