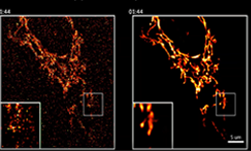
Ishtar Nyawira, a data science intern at the Pittsburgh Supercomputer Center shares how she is using deep learning and the GPU-accelerated Bridges supercomputer to automate the process of biological image annotation from high-resolution scanning electron microscope (SEM) imagery. The goal of the project is to ultimately understand how the neurons in our brains are wired.
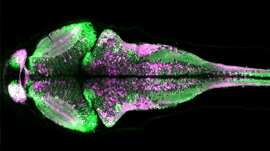
Harvard graduate students at the Allen Institute for Brain Science spent months manually annotating zebrafish neurons in SEM imagery. “The students were working with nearly 5,000 zebrafish images and each of them had to annotate the neurons in the images and there are about 200 neurons per image — so that takes a lot of time and incredibly time consuming,” said Nyawira. “This is something we’re trying to automate, so students don’t have to spend so much time doing things like that and can better apply their skills elsewhere.”
Using the Tesla P100 GPUs on the Bridges supercomputer and the TensorFlow deep learning framework, Ishtar is training her models on SEM imagery of zebrafish larva and the mouse brain to recognize neurons without being confused about noise and the tissue inside of the images.
For more details about Ishtar’s research project, watch her presentation in the NVIDIA booth at SC 16.
Share your GPU-accelerated work with us at http://nvda.ws/2cpa2d4.
Watch more scientists and researchers share how accelerated computing is benefiting their work at http://nvda.ws/2dbscA7
Developer Spotlight: Automated Biomedical Image Annotation
Mar 24, 2017
Discuss (0)
Related resources
- GTC session: Learn how to Streamline Medical Imaging Annotation and Training workflows with MONAI Cloud APIs
- GTC session: MONAI Label: AI-Assisted Annotation for Continuous Learning for Radiology, Pathology, and Medical Video Data
- GTC session: Accelerating Development of Medical Imaging AI for BioPharma
- SDK: MONAI Label
- SDK: cuCIM
- Webinar: The Impact of Large Language Models (LLMs) on Life Sciences