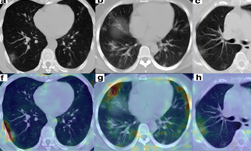
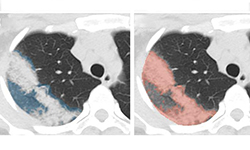
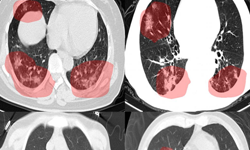
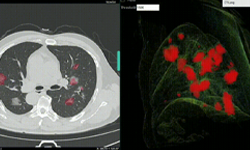
In a new paper published in Nature Communications, researchers at NVIDIA and the National Institutes of Health (NIH) demonstrate how they developed AI models (publicly available on NVIDIA NGC) to help researchers study COVID-19 in chest CT scans in an effort to develop new tools to better understand, measure and detect infections. Chest CT is emerging as a valuable diagnostic tool for clinical management of COVID-19.
This classification model for differentiation of pneumonia due to COVID-19 from other causes of pneumonia may find significant use during fall cold and flu season, to help differentiate COVID-19 from other viral, bacterial or fungal community-acquired pneumonias.
This model’s strength is that it was trained using a multinational, diverse, data set of more than 1,200 patients. COVID-19 CT scans were obtained from four hospitals across China, Italy, and Japan, where there was a wide variety in clinical timing and practice for CT use in outbreak settings. The generalizability of the model was evident when tested on an independent previously unseen demographic.
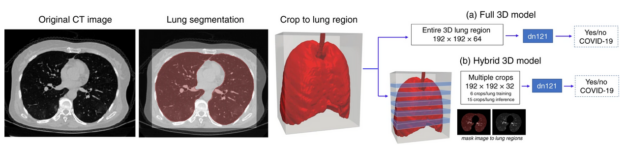
In total, 2,724 scans from 2,619 patients were used in this study, including 1,029 scans of 922 patients with RT-CPR confirmed COVID-19 and lung lesions related to COVID-19 pneumonia. Of these, 1,387 scans from 1,280 patients were utilized for algorithm development, and 1,337 patients were utilized for algorithm testing and evaluation.
This work encompassed two models that were used in series to come up with the COVID final classification model. The first model was a segmentation model that was used to define the lung regions which were subsequently used by the classification model. Initially, they developed two classification models — one utilizing the entire lung region with fixed input size (full 3D), and one utilizing average score of multiple regions within each lung at fixed image resolution (hybrid 3D).
Training converged at highest validation accuracy of 92.4% and 91.7% for hybrid 3D and full 3D classification models, respectively, for the task determining COVID-19 vs. other conditions. The validation accuracy in an unseen independent test set was observed with the 3D classification model (93.9%), with resultant probability of COVID-19 disease demonstrating 0.941 AUC. This was the model that was selected at the end, and published on NVIDIA NGC.
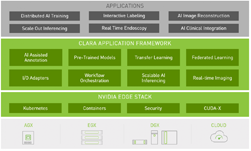
NGC is the hub for GPU-optimized AI, HPC, and data analytics software that can be quickly deployed on GPU-powered systems in the data center, at the edge, and in the cloud. NGC also hosts AI models and industry-specific SDKs like Clara that simplify building and deploying AI applications.
To assess the utility of these models for COVID-19 sensitivity at independent institutions, the cohort of COVID-19 patients from Tokyo, Japan were removed from training and validation datasets and models were re-trained utilizing identical algorithm configuration and hyperparameters as the original models. Overall, validation and testing accuracy were stable between models trained with and without patients from the leave-out institution.
The AI models were developed using the NVIDIA Clara application framework for medical imaging. NVIDIA Clara contains domain-specific AI training and deployment workflow tools that allowed NVIDIA and NIH to develop the models in under three weeks.
The fact that models were able to distinguish between COVID-19 and other types of pneumonia illustrates there may be a role for AI as one element of a CT-enhanced diagnosis, which could be immediately available. Subsequent models may include resource allocation, point of care detection for isolation of asymptomatic patients, prognostication, or monitoring for response in clinical trials for medical countermeasures such as anti-viral drugs or monoclonal antibodies, or disease severity during vaccine trials.